| Prevention of adriamycin-induced mdrl gene amplification and expression |

|

|

|
Page 4 of 7
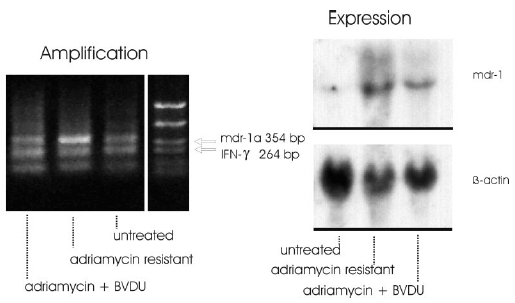
Figure 2: As result of these experiments it can be stated that the signal intensity of the mdr1a specific band appeared markedly increased in adriamycin resistant cells. The signal intensities of the adriamycin + BVDU treated cells resembled the intensity ratio of the untreated control cells (Fig. 2 left).
The expression of the mdr1-gene was studied in adriamycin-sensitive and in adriamycin-resistant F4-6 cells selected using Northern blot analysis. Levels of b-actin mRNA were also analyzed comparatively. b-Actin was used as an internal control for equivalent amounts of RNA loaded. Total cellular RNA from drug-sensitive and -resistant F4-6 cells was isolated by the method of Chomczynski & Sacchi (1987). 30 µg RNA was denatured by glyoxalation, electrophoresed through a 1% agarose gel, and transferred to Biodine A membrane (PALL, Portsmouth, UK) by the capillary blotting technique using 20 x SSD solution (1 x SSD contains 0.15 M sodium chloride and 0.015 M sodium citrate). RNA was immobilized by baking the membrane at 80° C for 1 h. DNA probes for hybridization were labeled with [a-32P]-dCTP by the random primer method using the Multiprime DNA labeling system from Amersham (Braunschweig, FRG). Membrane hybridization was carried out after 6 h prehybridization as described by Braun et al. (1989). Hybridized blots were washed in 2x SSD and 0.1 % SDS at room temperature for 20 min and at 50° C for 60 min, in 0.5 x SSC and 0.1 % SDS at 50° C, and finally in 0.2 x SSC and 0.1 % SDS at 55° C for 30 min. The membranes were exposed at –80° C to an X-ray film (Fuji Photo Film, Tokyo/Japan) using intensifying screens. The membranes were sequentially hybridized with the DNA probes. The following probes were used for hybridization: human mdr1 probe pHDR5A, 1.38 kb, and human b-actin cDNA probe, 1.2 kb.
|
|||||||||